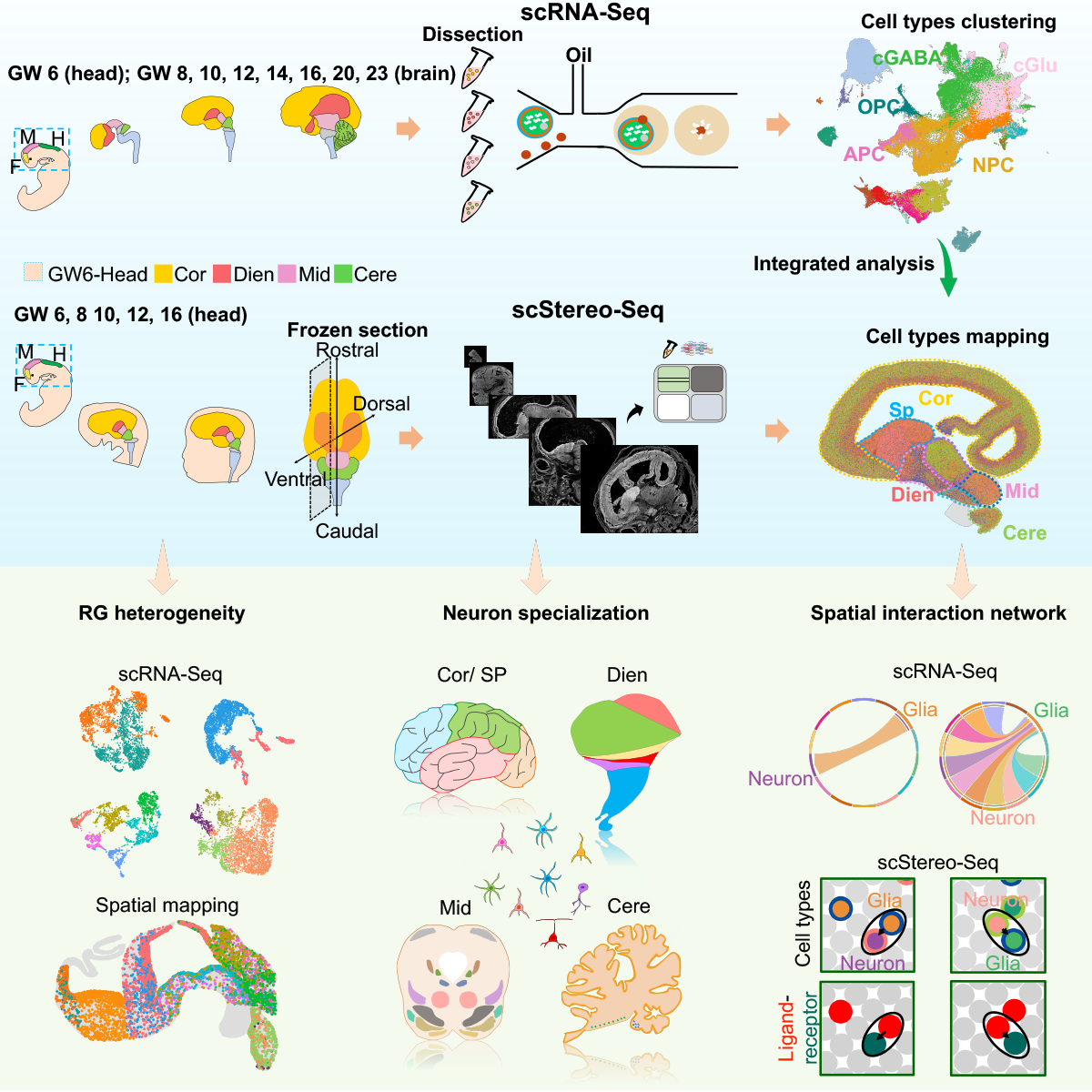
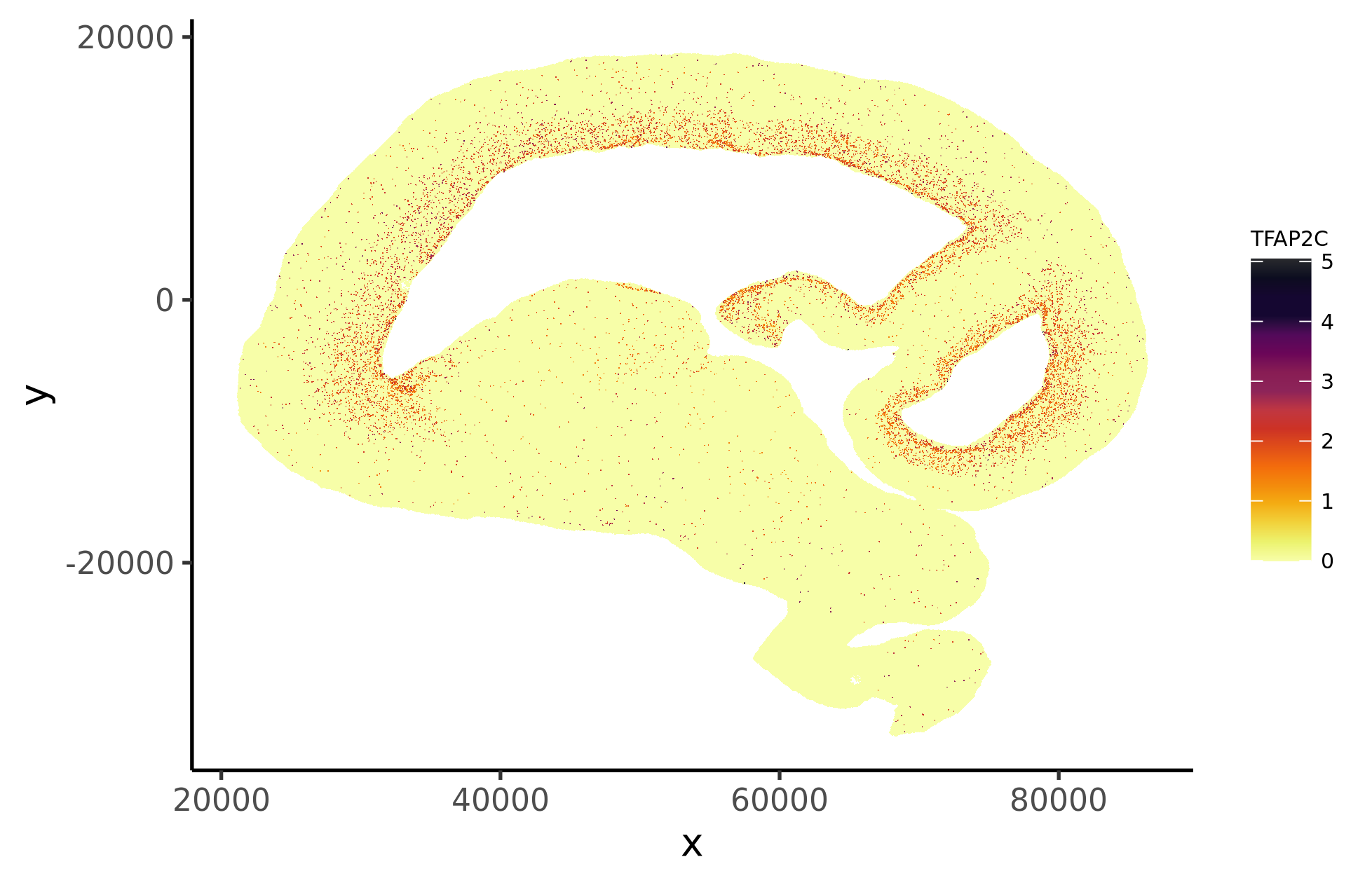
Different functional regions of brain are fundamental for basic neurophysiological activities. However, the regional specification remains largely unexplored during human brain development. Here, by combining spatial transcriptomics (scStereo-seq) and scRNA-seq, we built the first spatiotemporal developmental atlas of multiple human brain regions from 6-23 gestational weeks (GWs). We discovered that, around GW8, radial glia (RG) cells have displayed regional heterogeneity and specific spatial distribution. Interestingly, we found that the regional heterogeneity of RG subtypes contributed to the subsequent neuronal specification. Specifically, two diencephalon-specific subtypes gave rise to glutamatergic and GABAergic neurons, while subtypes in ventral midbrain were associated with the dopaminergic neurons. Similar GABAergic neuronal subtypes were shared between neocortex and diencephalon. Additionally, we revealed that cell-cell interactions between oligodendrocyte precursor cells and GABAergic neurons influenced and promoted neuronal development coupled with regional specification. Altogether, this study provides comprehensive insights into the regional specification in the developing human brain.
1. Spatiotemporal developmental atlas of distinct human brain regions is built.
2. Heterogeneous RG subtypes with specific spatial distribution are revealed.
3. RG regional heterogeneity contributes to the regional specification of neurons.
4. Interactions between OPCs and GABAergic neurons promote regional specification.